A Computational Model of the Cytoskeletal Network of Endothelial Cell Layers
The mechanical state of deformation as well as forces inside cells and at their boundaries play an important role for cell fate. Processes such as cell movement on a substrate, proliferation and apoptosis are strongly affected by mechanical cues. The mechanisms by which these processes are triggered, transferred and regulated are referred to as mechanosensing and mechanotransduction.
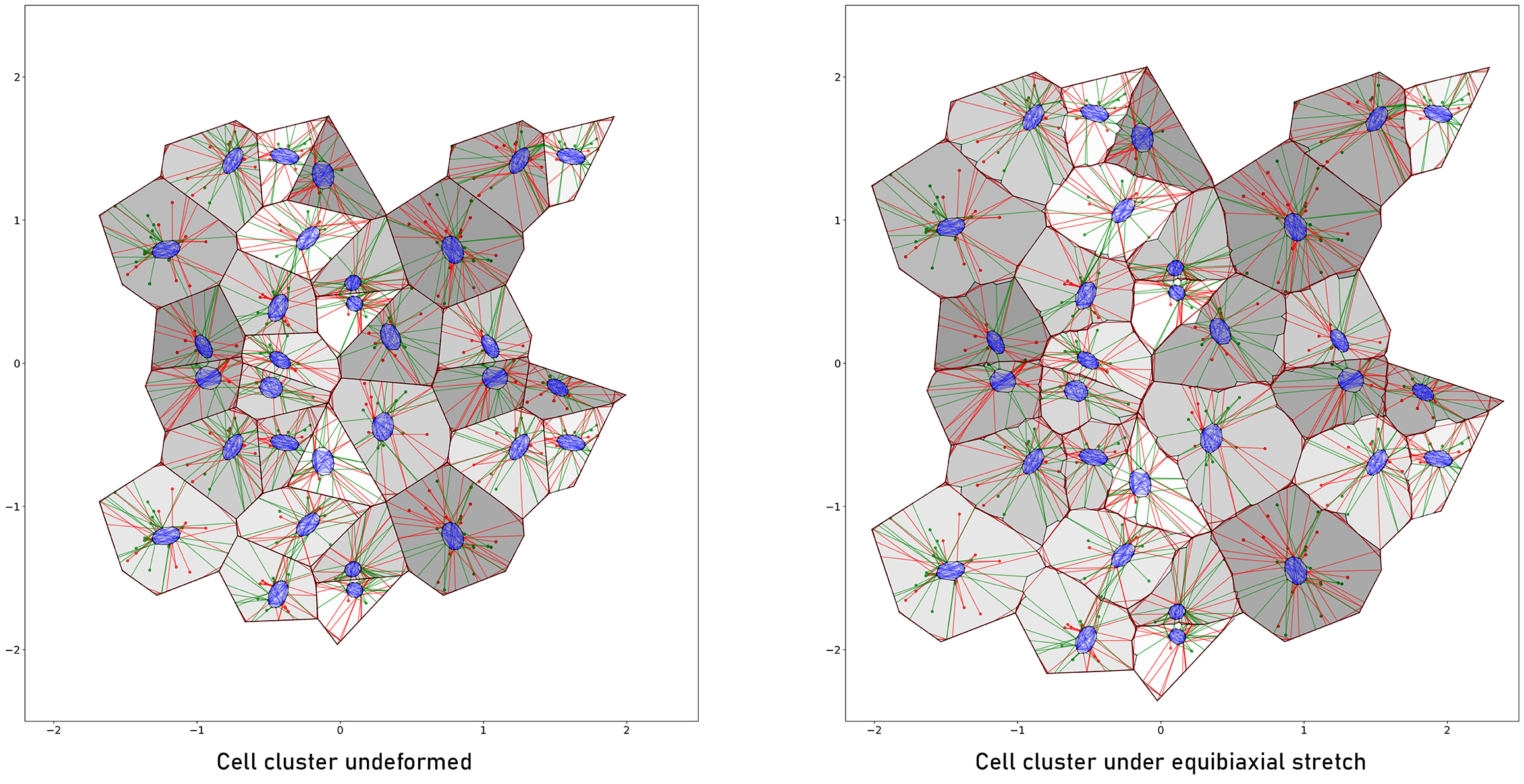
Computational models of cells and cell layers, e.g. [1, 2], can help understanding the complex interactions within the cells and in between them, as well as at the interface with substrate materials. This project is dedicated to endothelial mechanobiology and focusses on a new computational approach specific to the interlinked cytoskeletal networks of endothelial cell layers.
Through computational analysis we aim to understand the short-term deformation patterns in the endothelium when subjected to passive loads through the substrate material, and to rationalize the long-term adaptive mechanobiological response of the cells.
To this end we create computational models of human endothelial cell layers, including actin fibers, intermediate filaments and microtubules, which represent the key cytoskeletal components, as well as their connections over the cell boundaries. Laboratory experiments on endothelial cell cultures serve to inform and validate their digital twins through in-situ testing, traction force microscopy, and digital image analysis.
[1] Wang, L., & Chen, W. (2019). Modelling cell origami via a tensegrity model of the cytoskeleton in adherent cells. Applied Bionics and Biomechanics, 2019. external page https://doi.org/10.1155/2019/8541303
[2] Mosaffa, P., Rodríguez-Ferran, A., & Muñoz, J. J. (2018). Hybrid cell-centred/vertex model for multicellular systems with equilibrium-preserving remodelling. International Journal for Numerical Methods in Biomedical Engineering, 34(3). external page https://doi.org/10.1002/cnm.2928
